Microbial metabolomics
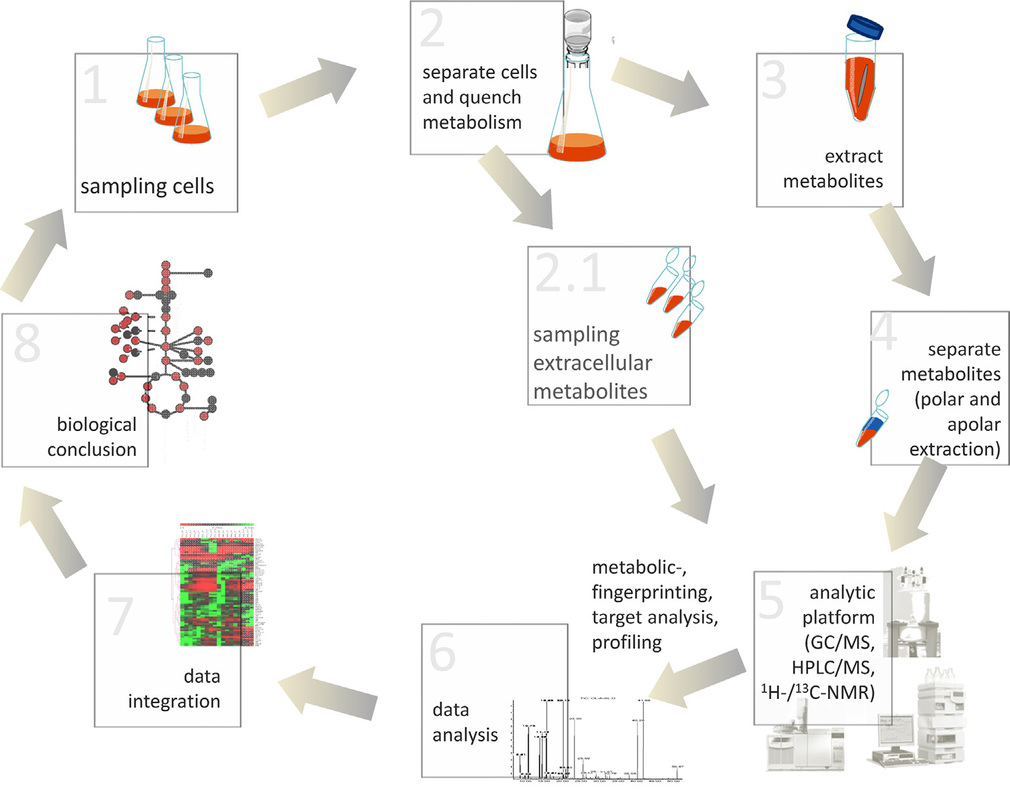
The field of metabolomics has become increasingly important in the context of functional genomics. Together with other ”omics“ data, the investigation of the metabolome is an essential part of systems biology. Beside the analysis of human and animal biofluids, the investigation of the microbial physiology by methods of metabolomics has gained increased attention. For example, the analysis of metabolic processes during growth or virulence factor expression is crucially important to understand pathogenesis of bacteria.
Common bioanalytical techniques for metabolome analysis include liquid and gas chromatographic methods coupled to mass spectrometry (LC-MS and GC-MS) and spectroscopic approaches such as NMR. In order to achieve metabolome data representing the physiological status of a microorganism, well-verified protocols for sampling and analysis are necessary.
Our current setup in lab consist of an Agilent GC-MS (EI) which allows us to measure more than 100 metabolites from very minute samples. This includes organic acids, amino acids, fatty acids, sugars, sterols etc.
We recently upgraded the lab to have a LC-MS/MS for high-throughput metabolomics available which covers depending on column installed hundreds of metabolites ranging from natural products, lipids, co-factors etc.
The field of metabolomics has become increasingly important in the context of functional genomics. Together with other ”omics“ data, the investigation of the metabolome is an essential part of systems biology. Beside the analysis of human and animal biofluids, the investigation of the microbial physiology by methods of metabolomics has gained increased attention. For example, the analysis of metabolic processes during growth or virulence factor expression is crucially important to understand pathogenesis of bacteria.
Common bioanalytical techniques for metabolome analysis include liquid and gas chromatographic methods coupled to mass spectrometry (LC-MS and GC-MS) and spectroscopic approaches such as NMR. In order to achieve metabolome data representing the physiological status of a microorganism, well-verified protocols for sampling and analysis are necessary.
- Protocol for Staphylococcus aureus metabolome analysis
- Protocol for Bacillus subtilis cell sampling - published by Hanna Meyer Michael Lalk's lab
Our current setup in lab consist of an Agilent GC-MS (EI) which allows us to measure more than 100 metabolites from very minute samples. This includes organic acids, amino acids, fatty acids, sugars, sterols etc.
We recently upgraded the lab to have a LC-MS/MS for high-throughput metabolomics available which covers depending on column installed hundreds of metabolites ranging from natural products, lipids, co-factors etc.